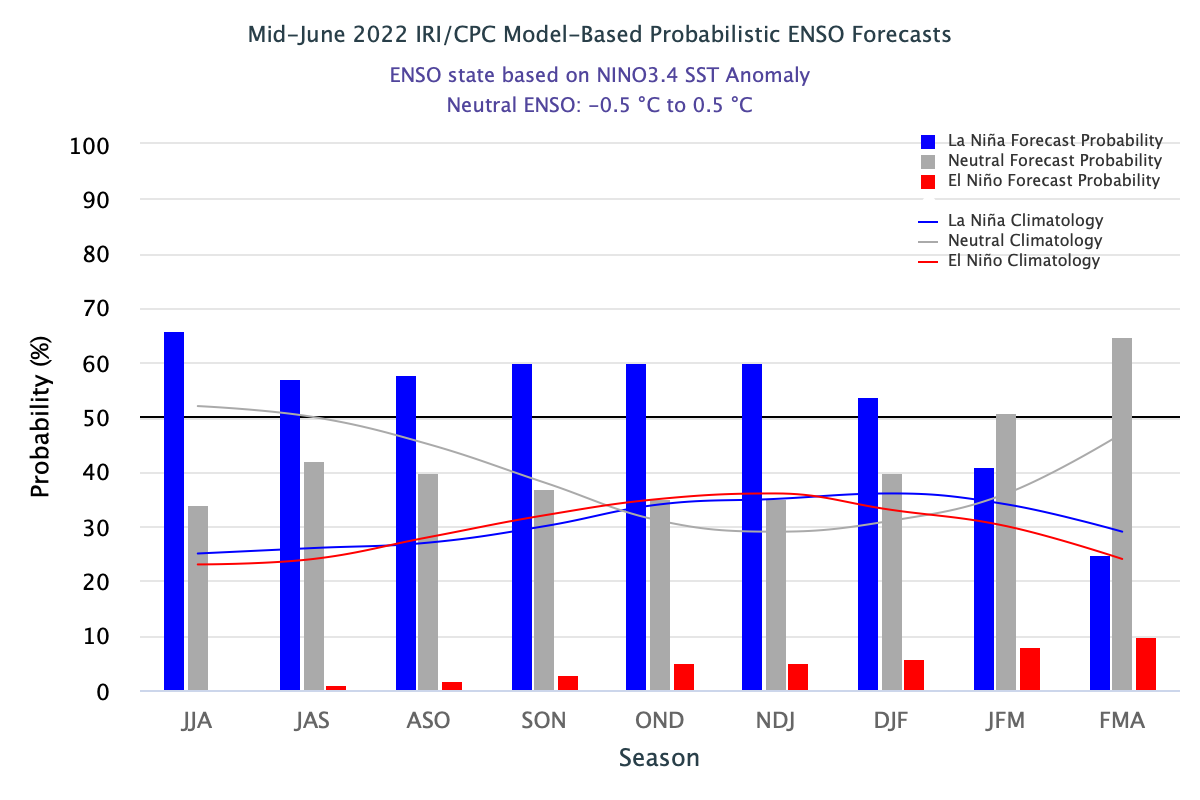
The ENSO predictions plume dataset contains the forecasted SST anomalies from various statistical and dynamical models from a range of third-party international forecasters. They range from weather agencies, government bodies to universities. Forecasted values start from Jan 2004 up until present. The dataset is compiled by the IRI from Columbia Climate School.
These models will provide monthly forecasts for up to nine overlapping 3-month periods. The 3-month periods are abbreviated by the starting letter of the month. For example, the 3-month period of Jan-Feb-Mar will be referred to as JFM.
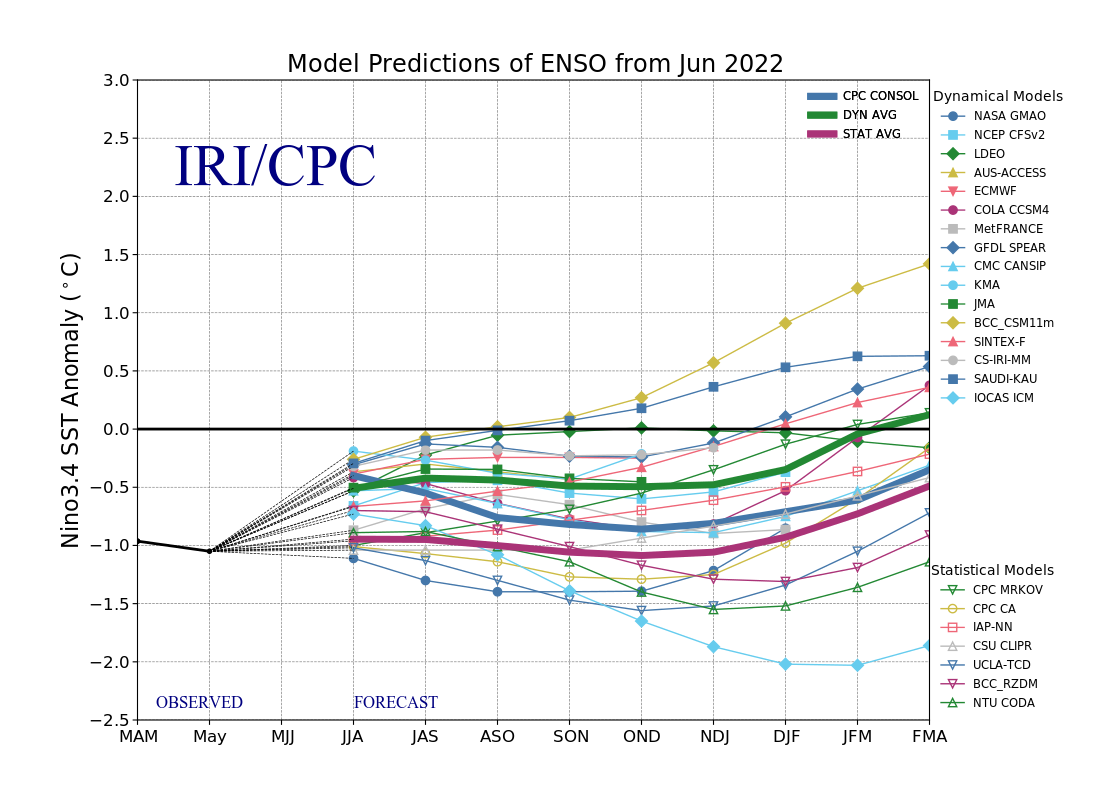
For the simplicity of our research, we will convert the interpretation of the 3 month period into corresponding stand-alone months. For instance, in our subsequent visualisations, forecasted values for the 3-month period of Jan-Feb-Mar will be tagged to March (the last month of the 3-month period). This will be further observed later when we discuss the results of our visualisations. This will make it easier for us to plot time values instead of having to deal with 3-month periods on the x-axis.
On the 19th of each month, the IRI will release the updated forecasted values for the given month on its website.
To investigate the accuracy of the forecasted values, we will aggregate all values and take the average from all models. We will extract and transform the data from enso_plumes.json, after which visualize the trends and accuracy of the forecasts.